Multimodal RAG with Colpali, Milvus and VLMs
In this post, we will see how to do multimodal RAG with colpali, milvus and a visual language model (gemini/gpt-4o).
We will build an application to upload a PDF and then do Q&A queries on it. Q&A can be done on both text and visual elements of the PDF. We will not extract text from the PDF; instead, we will treat it as an image and use colpali to get embeddings for the PDF pages. These embeddings will be indexed to Milvus, and then we will use a VLM to do Q&A queries on the PDF pages.
If you just want to see the code in action, there is a demo at https://huggingface.co/spaces/saumitras/colpali-milvus. Code for the same is here.
TOC:
- Problem
- Why colpali?
- Understanding how colpali works
- Code to upload a PDF, get embedding using colpali, index it to Milvus, then do Q&A queries using a vision language model (gemini/openai)
Problem
Let's say a company wants to build a Q&A/search interface for its internal documents, which include PDFs, word files, wikis, images, and text files. The traditional approach involves extracting text and media, detecting layout for structure, and indexing the information in a vector store for semantic search. However, this method often falls short for complex documents containing images, tables, and graphs. Let's look at an example below:
We have a PDF with stats on covid in the form of charts and tables. We want to answer the queries below:
1. What is the correlation between the samples tested and the positivity rate?
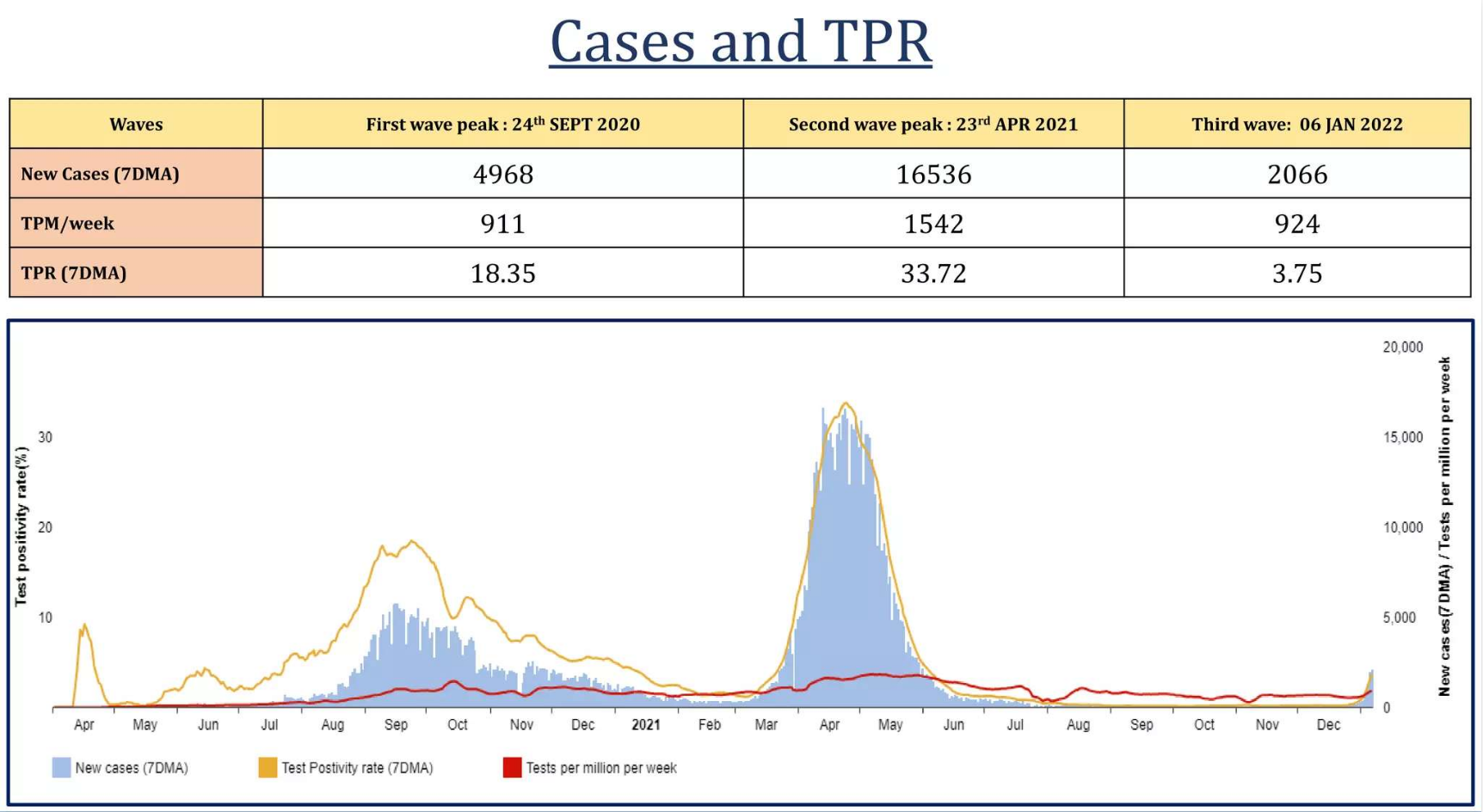
2. When and what was the highest number of cases and TPR?
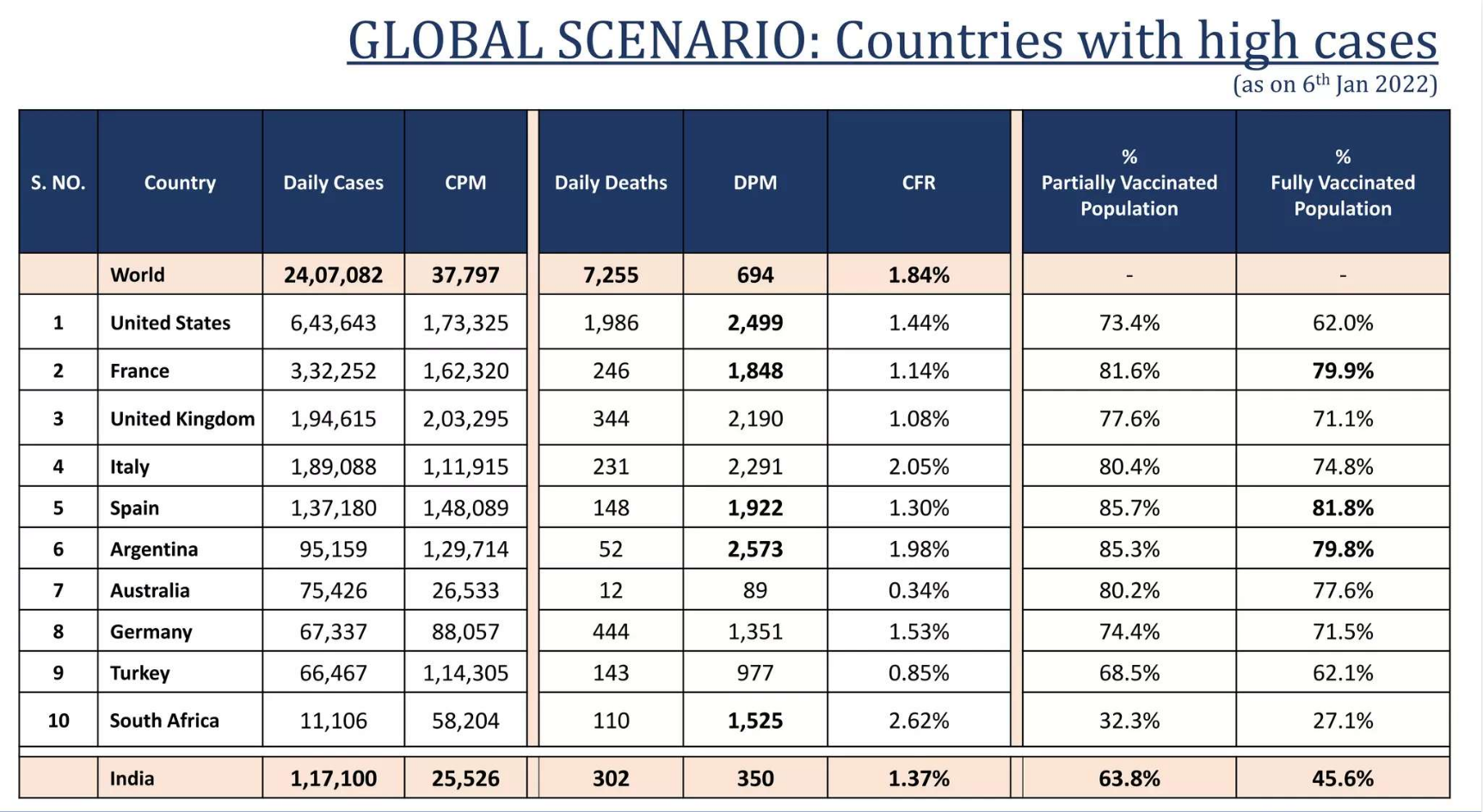
3. Which country had the highest omicron cases?
These queries can be answered by using data from following 3 pages:
Page 4: A chart showing stats on samples and positivity rate
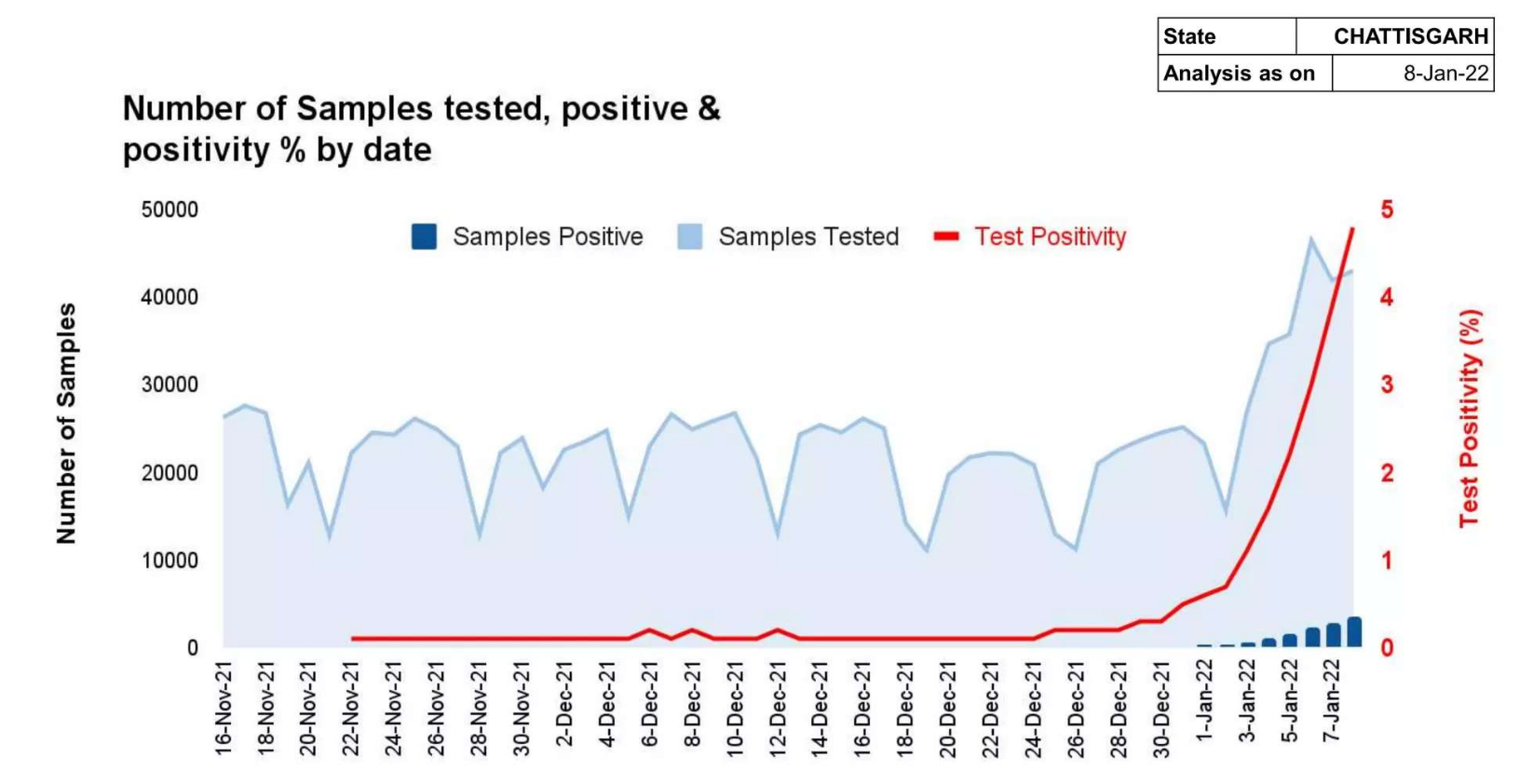
Page 8: A table showing cases and TPR
Page 9: A table showing cases by country
It would be difficult to extract data from these pages as text in a manner which can be used for querying. We want to show user the answer and source page(s) from the PDF which contains the answer, like below:
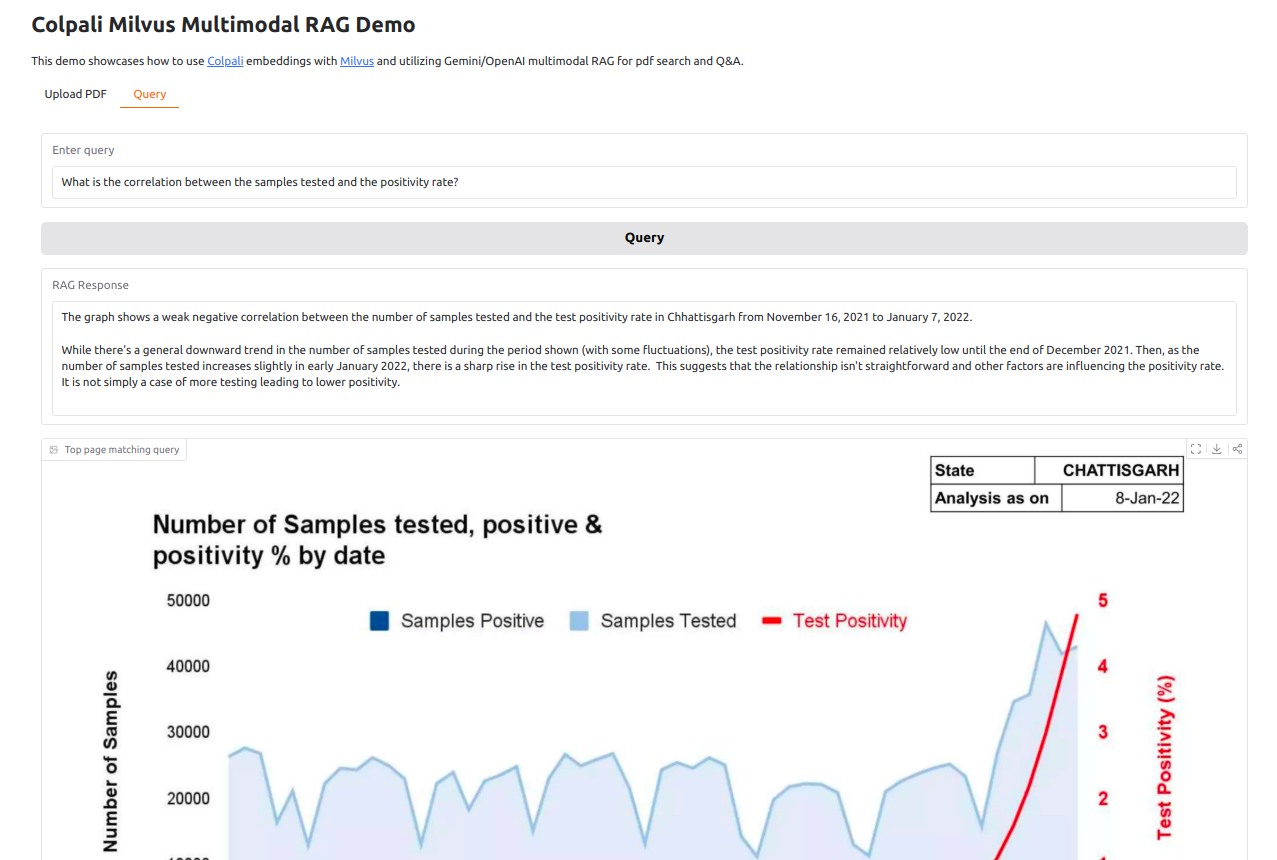
Let's understand how colpali can help us here.
Why colpali?
Document retrieval has always been a key component of systems like search engines and information retrieval. Traditional document retrieval methods rely heavily on text-based methods (like OCR and text segmentation), often missing crucial visual cues like layouts, images, and tables.
Colpali addresses this by using Vision-Language Models (VLMs) to understand and retrieve visually rich documents, capturing both textual and visual information. Colpali's architecture allows direct encoding of document images into a common embedding space, eliminating the need for time-consuming text extraction and segmentation.
Understanding how colpali works
Colpali works in the following steps:
Step 1: Treating the Document as an Image
Imagine we have a PDF document. Normally, we would extract text from the document using OCR (Optical Character Recognition), segment it into different sections, and then use these segments for searching. colpali simplifies this process by treating the entire document page as an image, bypassing the need for complex text extraction, layout detection, or OCR.
Step 2: Splitting the Image into Patches
Once colpali has this "image" of the document, it divides the page into small, uniform pieces called patches. Each patch captures a tiny portion of the page. It might contain a few words, a piece of a graph, or part of an image. This division helps the model focus on the document's small, detailed parts rather than trying to understand the whole page at once.
At first glance, it might seem like dividing an image into patches is similar to breaking text into chunks. However, these two methods have several key differences, especially in how they handle and preserve context. Let’s dive deeper into these differences to understand why patch-based processing in colpali is more effective for document retrieval compared to traditional text chunking.
Understanding Context Loss in Text Chunking
In traditional text chunking, text is split into smaller chunks based on certain tokens since many models limit the number of tokens they can process at once.
Problem with Context Loss:
- Chunking can split sentences or paragraphs midway, causing crucial context to be lost. It can also result in incomplete information in one chunk and missing context in another. Chunking doesn't preserve visual or structural information, such as the relationship between headings and their corresponding content or the placement of text in tables or figures.
For example, If you have a document with a heading followed by a table, text chunking might separate the heading and the table, losing the context that the table belongs to that heading.
Patch-Based Image Processing in colpali
Colpali divides the document image into patches, much like dividing a photo into small squares. Each patch is a fixed-size portion of the image, like a mini-snapshot of that part of the page.
Patches are more effective due to the following reasons:
- No Loss of Structure: The patches retain the document's visual structure, preserving its spatial layout. For instance, if a page has two columns of text or a table with rows and columns, each patch maintains its relative position, ensuring that the model understands the overall arrangement of the elements.
- Multi-Modal Context: Patches capture both textual and visual information. This includes both visual features (e.g., font styles, colors, boldness) and non-text elements (e.g., figures and graphs).
- Positional Awareness: Each patch has a positional embedding that tells the model where it is located on the page, helping the model understand the overall layout.
Step 3: Embedding Creation and Aligning Visual and Textual Information
Each patch is then passed through a Vision Transformer (ViT), which converts them into unique embeddings. Next, colpali aligns these visual embeddings with the text of the query by transforming the query into its own set of embeddings. colpali uses a process called alignment that aligns image path embeddings and text embeddings in the same vector space. Only then can we compare the similarity between query and document embeddings.
Step 4: Scoring the Relevance - Late Interaction Mechanism
At this point, colpali has embeddings for both the query and the document. The next challenge is to identify the relevant parts of the document. colpali uses a process called the Late Interaction Mechanism, where each piece of the query is finely matched against every part of the document, scoring and ranking their relevance.
Colpali highlights the most relevant pieces of the document, focusing on the patches that best match the query. This approach enables colpali to efficiently retrieve relevant information from visually rich documents, capturing both visual and textual data without losing context.
Code
Full code at https://github.com/saumitras/colpali-milvus-rag/
1. Add colpali processor
model_name = "vidore/colpali-v1.2"
device = get_torch_device("cuda")
model = colpali.from_pretrained(
model_name,
torch_dtype=torch.bfloat16,
device_map=device,
).eval()
processor = cast(colpaliProcessor, colpaliProcessor.from_pretrained(model_name))
2. Use colpali to get embeddings for image (pdf pages)
def process_images(self, image_paths:list[str], batch_size=5):
print(f"Processing {len(image_paths)} image_paths")
images = self.get_images(image_paths)
dataloader = DataLoader(
dataset=ListDataset[str](images),
batch_size=batch_size,
shuffle=False,
collate_fn=lambda x: processor.process_images(x),
)
ds: List[torch.Tensor] = []
for batch_doc in tqdm(dataloader):
with torch.no_grad():
batch_doc = {k: v.to(model.device) for k, v in batch_doc.items()}
embeddings_doc = model(**batch_doc)
ds.extend(list(torch.unbind(embeddings_doc.to(device))))
ds_np = [d.float().cpu().numpy() for d in ds]
return ds_np
3. Use colpali to get embeddings for text (user query)
def process_text(self, texts: list[str]):
print(f"Processing {len(texts)} texts")
dataloader = DataLoader(
dataset=ListDataset[str](texts),
batch_size=1,
shuffle=False,
collate_fn=lambda x: processor.process_queries(x),
)
qs: List[torch.Tensor] = []
for batch_query in dataloader:
with torch.no_grad():
batch_query = {k: v.to(model.device) for k, v in batch_query.items()}
embeddings_query = model(**batch_query)
qs.extend(list(torch.unbind(embeddings_query.to(device))))
qs_np = [q.float().cpu().numpy() for q in qs]
return qs_np
4. Code to create collection, index and query in milvus
class MilvusManager:
def __init__(self, milvus_uri, collection_name, create_collection, dim=128):
self.client = MilvusClient(uri=milvus_uri)
self.collection_name = collection_name
if self.client.has_collection(collection_name=self.collection_name):
self.client.load_collection(collection_name)
self.dim = dim
if create_collection:
self.create_collection()
self.create_index()
def create_collection(self):
if self.client.has_collection(collection_name=self.collection_name):
self.client.drop_collection(collection_name=self.collection_name)
schema = self.client.create_schema(
auto_id=True,
enable_dynamic_fields=True,
)
schema.add_field(field_name="pk", datatype=DataType.INT64, is_primary=True)
schema.add_field(
field_name="vector", datatype=DataType.FLOAT_VECTOR, dim=self.dim
)
schema.add_field(field_name="seq_id", datatype=DataType.INT16)
schema.add_field(field_name="doc_id", datatype=DataType.INT64)
schema.add_field(field_name="doc", datatype=DataType.VARCHAR, max_length=65535)
self.client.create_collection(
collection_name=self.collection_name, schema=schema
)
def create_index(self):
self.client.release_collection(collection_name=self.collection_name)
self.client.drop_index(
collection_name=self.collection_name, index_name="vector"
)
index_params = self.client.prepare_index_params()
index_params.add_index(
field_name="vector",
index_name="vector_index",
index_type="HNSW",
metric_type="IP",
params={
"M": 16,
"efConstruction": 500,
},
)
self.client.create_index(
collection_name=self.collection_name, index_params=index_params, sync=True
)
def create_scalar_index(self):
self.client.release_collection(collection_name=self.collection_name)
index_params = self.client.prepare_index_params()
index_params.add_index(
field_name="doc_id",
index_name="int32_index",
index_type="INVERTED",
)
self.client.create_index(
collection_name=self.collection_name, index_params=index_params, sync=True
)
def search(self, data, topk):
search_params = {"metric_type": "IP", "params": {}}
results = self.client.search(
self.collection_name,
data,
limit=int(50),
output_fields=["vector", "seq_id", "doc_id"],
search_params=search_params,
)
doc_ids = set()
for r_id in range(len(results)):
for r in range(len(results[r_id])):
doc_ids.add(results[r_id][r]["entity"]["doc_id"])
scores = []
def rerank_single_doc(doc_id, data, client, collection_name):
doc_colbert_vecs = client.query(
collection_name=collection_name,
filter=f"doc_id in [{doc_id}, {doc_id + 1}]",
output_fields=["seq_id", "vector", "doc"],
limit=1000,
)
doc_vecs = np.vstack(
[doc_colbert_vecs[i]["vector"] for i in range(len(doc_colbert_vecs))]
)
score = np.dot(data, doc_vecs.T).max(1).sum()
return (score, doc_id)
with concurrent.futures.ThreadPoolExecutor(max_workers=300) as executor:
futures = {
executor.submit(
rerank_single_doc, doc_id, data, self.client, self.collection_name
): doc_id
for doc_id in doc_ids
}
for future in concurrent.futures.as_completed(futures):
score, doc_id = future.result()
scores.append((score, doc_id))
scores.sort(key=lambda x: x[0], reverse=True)
if len(scores) >= topk:
return scores[:topk]
else:
return scores
def insert(self, data):
colbert_vecs = [vec for vec in data["colbert_vecs"]]
seq_length = len(colbert_vecs)
doc_ids = [data["doc_id"] for i in range(seq_length)]
seq_ids = list(range(seq_length))
docs = [""] * seq_length
docs[0] = data["filepath"]
self.client.insert(
self.collection_name,
[
{
"vector": colbert_vecs[i],
"seq_id": seq_ids[i],
"doc_id": doc_ids[i],
"doc": docs[i],
}
for i in range(seq_length)
],
)
def get_images_as_doc(self, images_with_vectors:list):
images_data = []
for i in range(len(images_with_vectors)):
data = {
"colbert_vecs": images_with_vectors[i]["colbert_vecs"],
"doc_id": i,
"filepath": images_with_vectors[i]["filepath"],
}
images_data.append(data)
return images_data
def insert_images_data(self, image_data):
data = self.get_images_as_doc(image_data)
for i in range(len(data)):
self.insert(data[i])
5. Save pdf as individual images
class PdfManager:
def __init__(self):
pass
def clear_and_recreate_dir(self, output_folder):
print(f"Clearing output folder {output_folder}")
if os.path.exists(output_folder):
shutil.rmtree(output_folder)
os.makedirs(output_folder)
def save_images(self, id, pdf_path, max_pages, pages: list[int] = None) -> list[str]:
output_folder = f"pages/{id}/"
images = convert_from_path(pdf_path)
print(f"Saving images from {pdf_path} to {output_folder}. Max pages: {max_pages}")
self.clear_and_recreate_dir(output_folder)
num_page_processed = 0
for i, image in enumerate(images):
if max_pages and num_page_processed >= max_pages:
break
if pages and i not in pages:
continue
full_save_path = f"{output_folder}/page_{i + 1}.png"
image.save(full_save_path, "PNG")
num_page_processed += 1
return [f"{output_folder}/page_{i + 1}.png" for i in range(num_page_processed)]
6. Middleware to index and search Milvus for embeddings generated from colpali
class Middleware:
def __init__(self, id:str, create_collection=True):
hashed_id = hashlib.md5(id.encode()).hexdigest()[:8]
milvus_db_name = f"milvus_{hashed_id}.db"
self.milvus_manager = MilvusManager(milvus_db_name, "colpali", create_collection)
def index(self, pdf_path: str, id:str, max_pages: int, pages: list[int] = None):
print(f"Indexing {pdf_path}, id: {id}, max_pages: {max_pages}")
image_paths = pdf_manager.save_images(id, pdf_path, max_pages)
print(f"Saved {len(image_paths)} images")
colbert_vecs = colpali_manager.process_images(image_paths)
images_data = [{
"colbert_vecs": colbert_vecs[i],
"filepath": image_paths[i]
} for i in range(len(image_paths))]
print(f"Inserting {len(images_data)} images data to Milvus")
self.milvus_manager.insert_images_data(images_data)
print("Indexing completed")
return image_paths
def search(self, search_queries: list[str]):
print(f"Searching for {len(search_queries)} queries")
final_res = []
for query in search_queries:
print(f"Searching for query: {query}")
query_vec = colpali_manager.process_text([query])[0]
search_res = self.milvus_manager.search(query_vec, topk=1)
print(f"Search result: {search_res} for query: {query}")
final_res.append(search_res)
return final_res
7. Use Gemini or gpt-4o to do Q&A on pdf page(s) matching user query
class Rag:
def get_answer_from_gemini(self, query, imagePaths):
print(f"Querying Gemini for query={query}, imagePaths={imagePaths}")
try:
genai.configure(api_key=os.environ['GEMINI_API_KEY'])
model = genai.GenerativeModel('gemini-1.5-flash')
images = [Image.open(path) for path in imagePaths]
chat = model.start_chat()
response = chat.send_message([*images, query])
answer = response.text
print(answer)
return answer
except Exception as e:
print(f"An error occurred while querying Gemini: {e}")
return f"Error: {str(e)}"
def get_answer_from_openai(self, query, imagesPaths):
print(f"Querying OpenAI for query={query}, imagesPaths={imagesPaths}")
try:
payload = self.__get_openai_api_payload(query, imagesPaths)
headers = {
"Content-Type": "application/json",
"Authorization": f"Bearer {os.environ['OPENAI_API_KEY']}"
}
response = requests.post(
url="https://api.openai.com/v1/chat/completions",
headers=headers,
json=payload
)
response.raise_for_status() # Raise an HTTPError for bad responses
answer = response.json()["choices"][0]["message"]["content"]
print(answer)
return answer
except Exception as e:
print(f"An error occurred while querying OpenAI: {e}")
return None
def __get_openai_api_payload(self, query:str, imagesPaths:List[str]):
image_payload = []
for imagePath in imagesPaths:
base64_image = encode_image(imagePath)
image_payload.append({
"type": "image_url",
"image_url": {
"url": f"data:image/jpeg;base64,{base64_image}"
}
})
payload = {
"model": "gpt-4o",
"messages": [
{
"role": "user",
"content": [
{
"type": "text",
"text": query
},
*image_payload
]
}
],
"max_tokens": 1024
}
return payload
In the next post, we will understand the limitations of colpali and a workaround for them.
References
- https://milvus.io/docs/use_colpali_with_milvus.md
- https://arxiv.org/abs/2407.01449